Related Post
Bioinformatics No Longer “Black Box”: Chip Sequencing Services| Standardized Data Analysis
2026-01-19Chip Sequencing Services have moved from niche experiments to mainstream engines of discovery, reshaping how scientists understand genome regulation and disease. Barski and colleagues profiled extensive histone modifications in Cell (2007), ChIP-seq has accelerated the field. The ENCODE Consortium, including teams led by Michael Snyder, scaled transcription factor atlases, while Richard A. Young’s group used ChIP-seq to define pluripotency networks in stem cells. At Longlight Technology, we build on this legacy with Chip Sequencing Services integrated end-to-end with Standardized Data Analysis to boost reproducibility and convert ChIP-seq data into insights – removing the bioinformatics “black box” and enabling confident decisions.
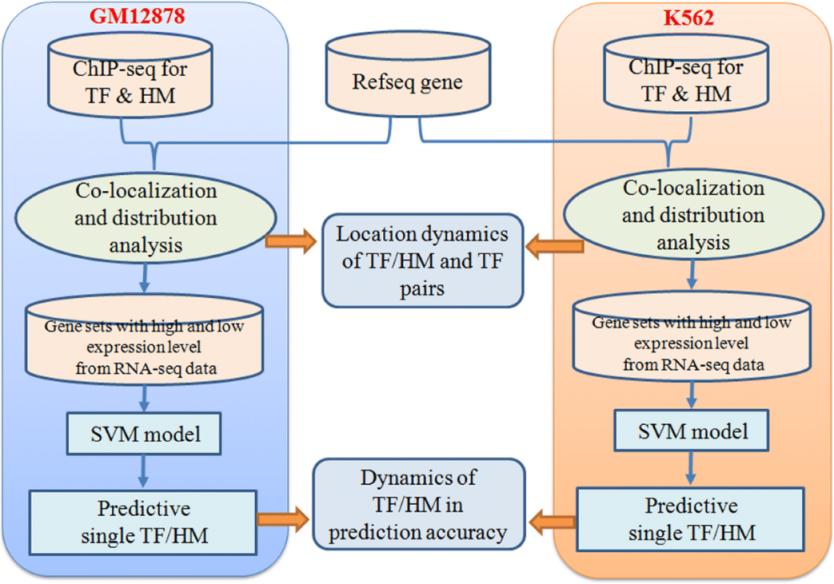
(Johnson, Mortazavi, Myers, Revealing transcription factor and histone modification co-localization
and dynamics across cell lines by integrating ChIP-seq and RNA-seq data, Science (2007))
What Chip Sequencing Services Deliver
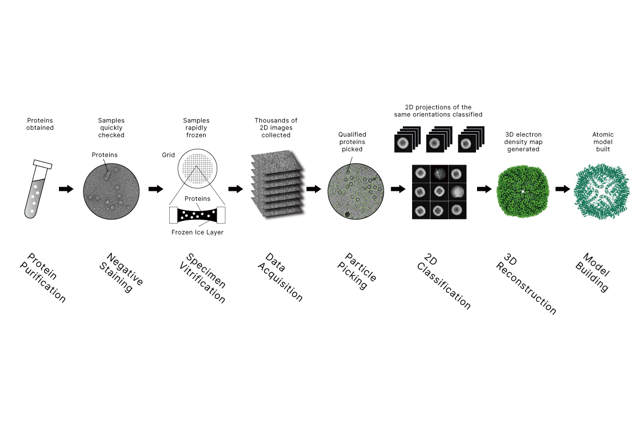
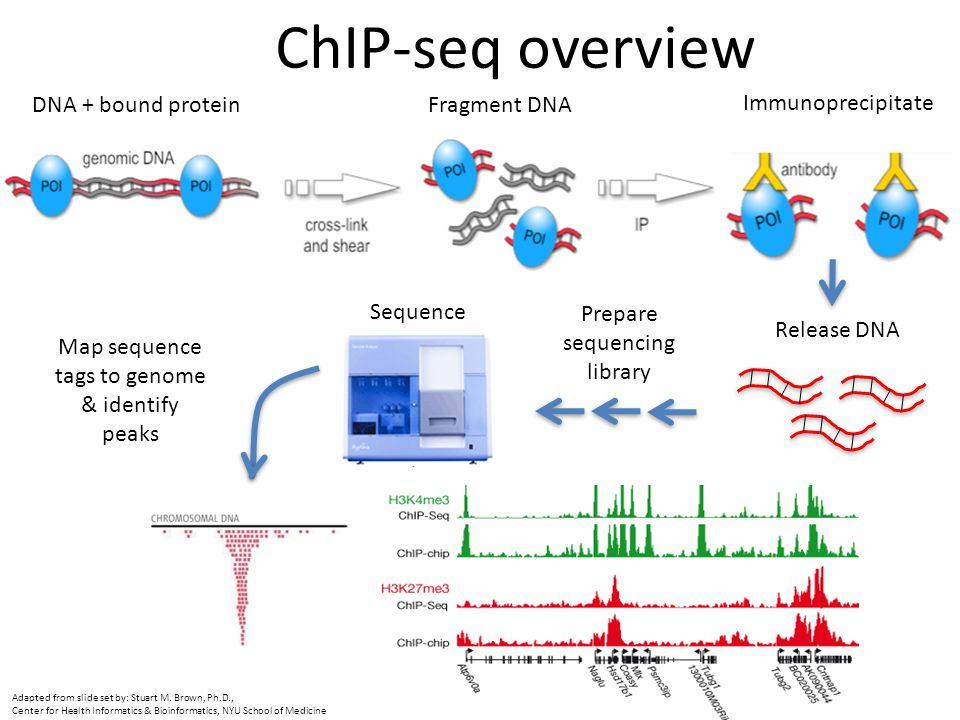
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) interrogates protein-DNA interactions across the genome. By capturing chromatin fragments bound by transcription factors or histones and reading them out with high-throughput sequencing, Chip Sequencing Services localize binding sites, map histone modification landscapes, and quantify occupancy changes under different conditions.
Longlight Technology provides a one-stop workflow: sample preparation, chromatin shearing, library construction, advanced sequencing, and rigorous data analysis. Our service is suitable for small sample sizes, supports analysis of specific genes or regions guided by your research purpose, and applies strict quality inspection at each link to ensure accuracy. Combined with our focused ultrasonication for precise chromatin shearing and high-quality reagents and consumables – including Qubit tubes and curated library preparation kits – we deliver complete data reports and raw data with transparent quality metrics.
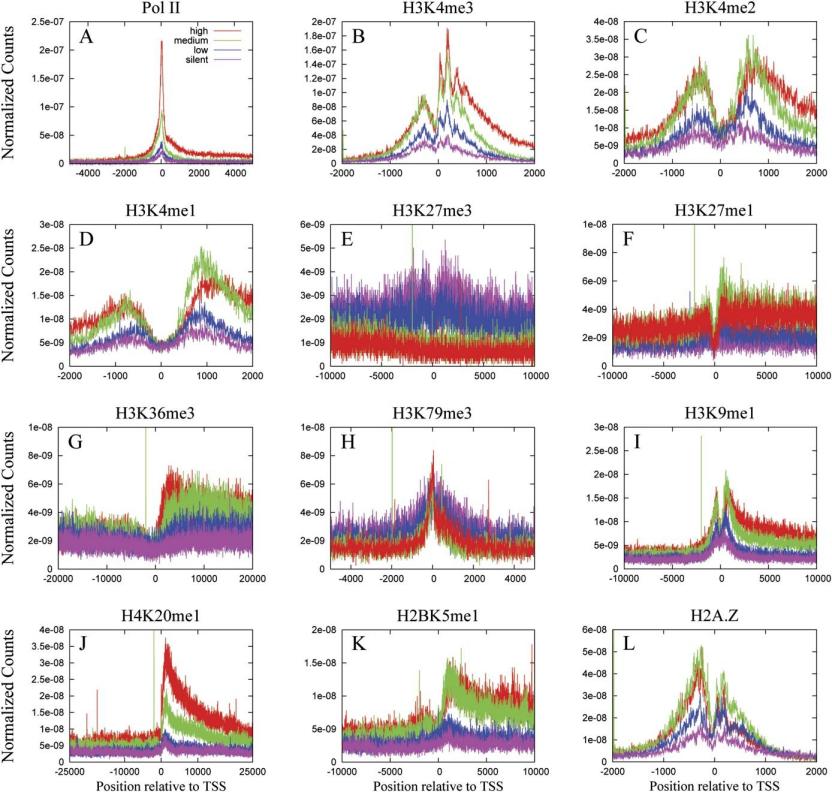
(High-Resolution Profiling of Histone Methylations in the Human Genome)
Bioinformatics No Longer a “Black Box“
The challenge with many Chip Sequencing Services is inconsistency: variable peak calling, non-comparable normalization, opaque QC, and fragmented deliverables. Longlight Technology enforces uniform standards across the analytical lifecycle, ensuring consistency from sample receipt to interpretive reporting.
Harmonized pipelines with validated controls and explainable, documented decisions underpin every run. QC matrix includes insert-size distribution, library complexity indices, alignment quality (e.g., mapping rate), duplication rate, background noise (e.g., FRiP/SNR), and reproducibility (e.g., replicate correlation/IDR). We deliver comprehensive reports that connect peaks to biological meaning – linking transcription factor occupancy with gene expression programs, histone marks with chromatin states, and co-binding events with regulatory modules. By enforcing common standards for inputs, processes, and outputs, we achieve cross-project comparability and prevent analytical drift. Bioinformatics becomes fully traceable, auditable, and decision-ready.
- Reproducibility By Design
• Reproducible engines: seeded, version-pinned tools, verified references, and parameter schemas per assay.
• Quantitative QC: instrument metrics, pre-align SNR/read quality, post-peak FRiP/NSC/RSC, replicate IDR.
• Complete package: raw/processed data, peak catalogs, annotated intervals, plus configuration and run-time provenance.
• Targeted analysis: concentrating on specific genes or regions when projects call for narrow, hypothesis-driven conclusions.
Why Longlight Technology
Longlight Technology integrates top-tier laboratory hardware with robust informatics to elevate Chip Sequencing Services across academic, clinical, and industrial settings.
• One-stop execution: Provide fixed cells or frozen tissue; we complete preparation, shearing, library building, sequencing, and bioinformatic analysis.
• Trustworthy by design: Continuous QC pinpoints the biology you care about – down to gene and region.
• Small sample? No problem. Our sensitivity-first approach extracts maximum insight.
• Reports that speak your language – TF binding, histone marks, co-binding patterns, and Pol II tracking.
• A unified consumables stack delivers consistency from gel to library.
• Focused ultrasonication: Precision shearing that increases enrichment specificity and improves library performance.
- From Sample to Insight: the Service Process
We simplify the move from a biological question to a confident answer. You submit fixed cells or frozen tissue; we return clear insights with minimal friction and complete transparency.
• Intake and standardized handling to maintain chromatin.
• Calibrated sonication for reproducible, target-fit fragments.
• Balanced ligation/PCR to preserve library complexity.
• Sequencing depth matched to TF/histone goals.
• Layered QC and alignment diagnostics.
• Peak/differential/motif/co-binding analyses with gene annotations.
• Reports, data, visuals, and interpretive summaries.
Applications and Impact
Chip Sequencing Services illuminate regulatory biology in contexts ranging from development to disease. By mapping the presence of proteins at genomic sites, you can draw protein occupancy maps within target regions, assay co-occurrence of histone marks in relation to gene expression, and position RNA polymerase II and trans-factor binding with precision. In pharmacology, ChIP-seq supports mechanism-of-action studies; in biopharmaceutical R&D, it validates target engagement; in academic research, it decodes transcriptional programs that govern cell fate.
Our approach connects ChIP-seq output to biological narratives. Motif analyses reveal sequence determinants of binding. Co-binding matrices highlight regulatory partnerships. Histone modification profiles segment chromatin states. Integrated annotations link peaks to promoters, enhancers, and gene bodies. Because the entire pipeline is standardized, longitudinal studies across time points, cell states, or treatments become directly comparable, reducing ambiguity and strengthening conclusions.
Call to Action
Ready for traceable pipelines and reproducible peaks? For TF, histone, or Pol II ChIP-seq, contact Longlight to book a consult, begin sample submission, or tour a sample report with QC metrics. We will help you convert ChIP-seq data into actionable insights with Standardized Data Analysis, end-to-end QC, and complete deliverables tailored to your research goals.